Dissecting the epigenome using CRISPR-based engineering technologies
The CRISPR-Cas9 revolution has fast-tracked our understanding of genetics and opened doors for curing many DNA-based diseases. This tool acts as molecular scissors that can be directed to specific sites of the genome when guided by a map-like molecule: the guide RNA. New technologies have adapted the CRISPR-Cas9 machinery to expand its use from a DNA-cutter capable of editing the genome, to a modulator that edits the epigenome. This offers potential to better understand how the epigenome influences cell types, and future possibilities for curing epigenetic diseases.
Repurposing CRISPR-Cas9 to edit the epigenome
To properly investigate the role of the epigenome in living organisms, scientists have mutated the molecular scissor Cas9—so that it can no longer cut DNA—and renamed it the deactivated, or dead Cas9 (dCas9). Because the core components of the CRISPR-dCas9 machinery remain intact, the tool can still recognize a guide RNA and be directed to a specific location of the genome.
Scientists are harnessing the dead enzyme’s function for epigenome editing by fusing it to epigenetic modifiers of their choice. Essentially, the histone or DNA methylation remodelers will hitch a ride on the dCas9, and the entire machinery will find its way to the location of interest by following the guide RNA that is custom-designed to target a specific genomic sequence. Once the guide RNA finds its target DNA sequence, the epigenetic modifier will remodel the epigenome at that spot using its normal function.
Mouse models engineered to express epigenome editors
Epigenome editing studies in living organisms are mainly done in mice. Because mice do not express a CRISPR-dCas9 epigenome editing system, scientists need to genetically integrate its coding sequence in safe-harbor sites—regions that will not disrupt normal genome function—of the mouse’s genome. A promoter must be placed in front of the coding sequence to allow it to be expressed; It can be constitutive (to express the epigenetic modifying machinery in all cell types) or cell type-specific or tissue-specific. To target the dCas9-epigenetic remodeler to specific stretches of DNA, the guide RNA can also be integrated into the mouse genome, or it can be delivered to cells using harmless viruses. These strategies provide a toolbox for scientists to answer many pressing questions in the field of epigenetics, in vivo.
How do epigenetic marks contribute to tissue-specific cell types?
Most cells that make up an individual are composed of the same DNA sequence. The epigenetic modifications that decorate the DNA influence how cells acquire their cell-type specificities. Regulatory loci such as promoters can be distinguished by their epigenetic signatures. For the most part, promoters that are turned on in specific cells are marked by high levels of activating H3K4me3 and/or H3K27ac and promoters that are turned off are marked by high levels of repressive H3K27me3 and/or H3K9me3. Studies probing epigenetic marks across the entire genome have been able to identify tissue-specific epigenetic signatures. For example, promoters of genes important for neuron activity will bear the active mark H3K4me3 in brain tissue but not in lung tissue. Nonetheless, these observations remain associative, and whether these epigenetic signatures causally turn genes on or off in a specific tissue is unknown.
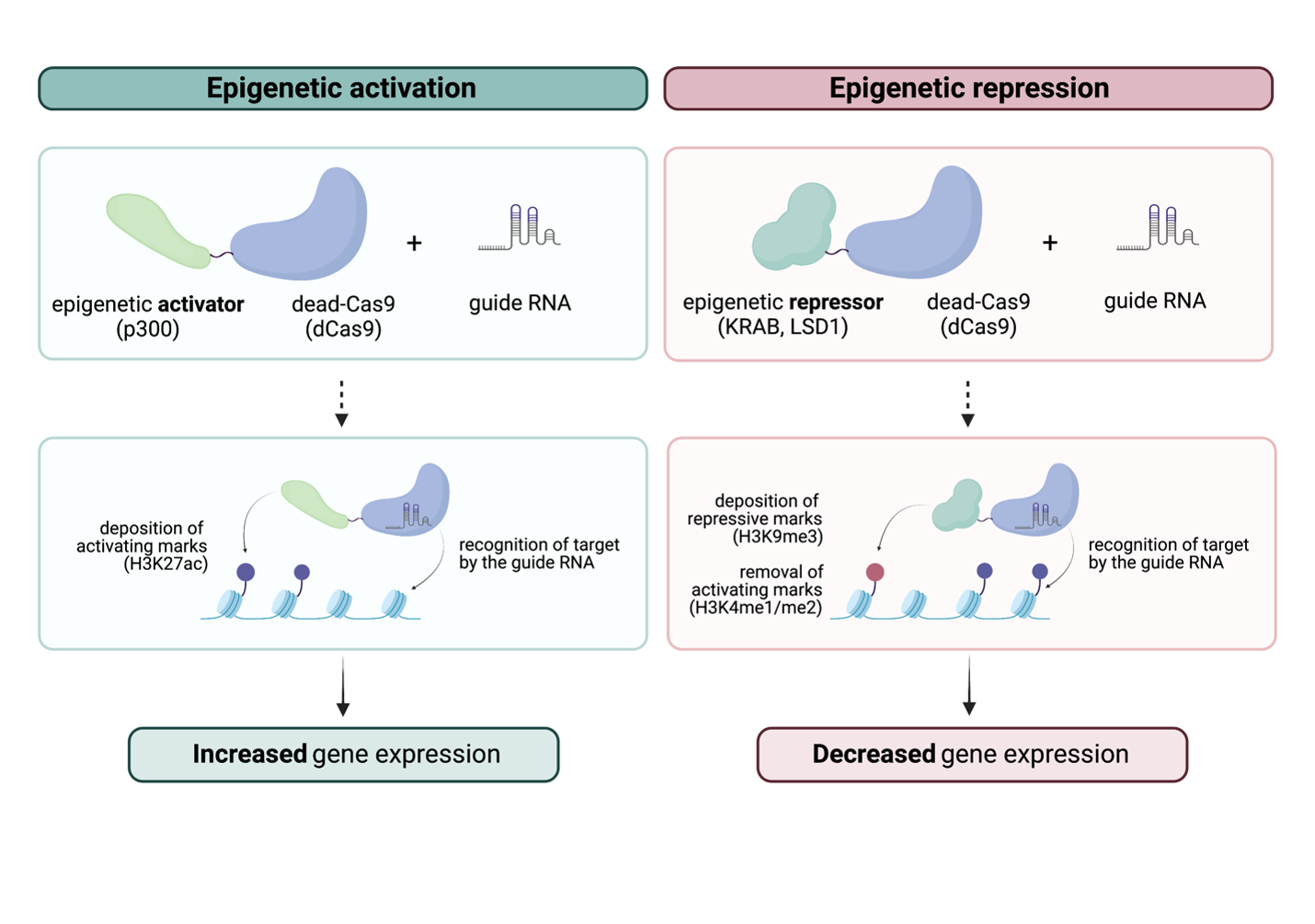
With epigenome editing engineering in mouse models, scientists can determine how epigenetic modifications at promoters from specific tissues influence gene expression. In one study, Gemberling and Siklenka et al. attached to the dCas9 either a KRAB transcriptional repressor, which promotes the deposition of repressive DNA methylation and H3K9me3 mark, or a transcriptional activator p300, which deposits the active H3K27ac mark. They genetically integrated the epigenome editors in the genome of two different mice strains to only be expressed specifically in the liver.
The team targeted the dCas9-p300 activator to Pdx1, a gene that can lead to early-onset diabetes when defective. They found that the dCas9-p300 tool was able to place H3K27ac specifically on the Pdx1 promoter and increase the gene’s expression. Still in the liver, they directed the dCas9-KRAB repressive machinery to Pcsk9, a gene involved in cholesterol reduction. Again, their targeting was highly specific, and they observed a decrease in Pcsk9 expression which was concordant with an increase in the repressive H3K9me3 mark at the Pcsk9 promoter. These findings indicate that the dCas9-p300 and dCas9-KRAB can robustly turn genes on and off in a tissue-specific manner.
How to decode the regulatory genome using epigenome engineering?
Enhancers are DNA sequences that fine-tune the transcription level of a gene located nearby or far away on the chromosome. Typically, multiple enhancers regulate the expression of a single gene, often in a tissue-specific manner. Additionally, enhancers are often redundant, meaning that removing one enhancer sequence from the genome does not always affect gene expression. Interestingly, like promoters, enhancers can be distinguished by their epigenetic signatures. Active enhancers generally bear high levels of H3K4me1 and H3K27ac, but low levels of H3K4me3.
Many groups have been taking advantage of these epigenome editing tools to target specific enhancers in the genome and see how the gene regulated by the enhancer reacts. Gemberling and Siklenka et al., aimed to activate one of the five enhancers flanking the Fos gene using their dCas9-p300 mouse model. By increasing H3K27ac levels at this specific enhancer, they were able to increase Fos gene expression in the brain.
In another study, Li et al., used a dual repressor system wherein mice expressed a dCas9-KRAB and dCas9-LSD1 to interrogate the function of numerous enhancers in the mouse genome. LSD1 is a histone demethylase that removes methyl groups from H3K4me1 and H3K4me2 and is known to shut down enhancer function. The authors probed enhancers that regulate genes involved in bone marrow formation (myelopoeisis) focusing on Cebpa, a gene implicated in leukemia, by targeting 4 enhancers flanking the gene. They found that 2 of the enhancers are necessary for Cebpa expression, one of which to a lesser extent than the other, while the other 2 were not required. These studies highlight the complexity of the regulatory genome and offer insight on how epigenome editing tools can be used to begin untangling it.
The tip of the iceberg
The new advances in epigenome editing technologies have geared scientists with tools to better analyze the epigenome. With hundreds of cell types and epigenetic modifications, and thousands of regulatory loci, gaining a more profound comprehension of our epigenome in a cell and tissue-specific manner is a daunting task. Nonetheless, our understanding of epigenetic regulation and the development of epigenome editing tools, is a first step towards manipulating the epigenome in the context of disease.
Learn More
- CRISPR technologies for precise epigenome editing: https://pubmed.ncbi.nlm.nih.gov/33420494/
- CRISPR 101: Epigenetics and editing the epigenome: https://blog.addgene.org/crispr-101-editing-the-epigenome
- Chromatin modifiers and remodelers: regulators of cellular differentiation: https://pubmed.ncbi.nlm.nih.gov/24366184/
We would like to thank Dr. Serge McGraw from Université de Montréal for his expert review of this article and Elizabeth Elder for providing a French translation.
Image created in www.BioRender.com