Epigenetic alteration of the young dark matter in sperm worsens fertility across two successive generations
Transposable elements, also known as “jumping genes”, are repetitive stretches of viral DNA that move from one position to another in the genome. Epigenetic inheritance studies have begun to shed light on the potential role of young transposable elements in transmitting epimutations from sperm to the next generation.
Epigenetic modifications keep transposable elements in check during embryo development.
In healthy adult cells, transposable elements are usually turned off by DNA methylation or repressive histone modifications. Loss of these epigenetic marks have been associated with cancer, schizophrenia, and other complex diseases.
During embryo development, most DNA methylation is erased in the embryo’s future sperm and egg cells. This DNA demethylation wave resets the DNA methylation of the embryo’s reproductive cells to a near-ground state. However, certain transposable elements need to remain methylated - or turned off - to avoid harm to the embryo and its future gametes.
Impairing DNA methylation in sperm leads to infertility in the next generation.
5,10-Methylenetetrahydrofolate reductase (MTHFR) is an important enzyme that helps generate methyl groups required for DNA methylation and histone methylation. In certain human populations, males with a MTHFR deficiency have abnormal DNA methylation patterns in their sperm and impaired fertility.
In a new study, Dr. Gurbet Karahan and Dr. Jacquetta Trasler from McGill University (Montreal), used mouse models to study how MTHFR deficiency affects DNA methylation in sperm and male fertility across two generations. They showed that a MTHFR deficiency in the first-generation fathers barely impacted their fertility. Interestingly, the MTHFR-deficient fathers gave rise to sons with abnormal reproductive functions.
The authors then questioned whether DNA methylation levels were altered in the sperm of the MTHFR-deficient fathers and their sons. They discovered a vast loss of DNA methylation in their sperm. Strikingly, over 80% of sites that lost DNA methylation in MTHFR fathers, also lost DNA methylation in their sons.
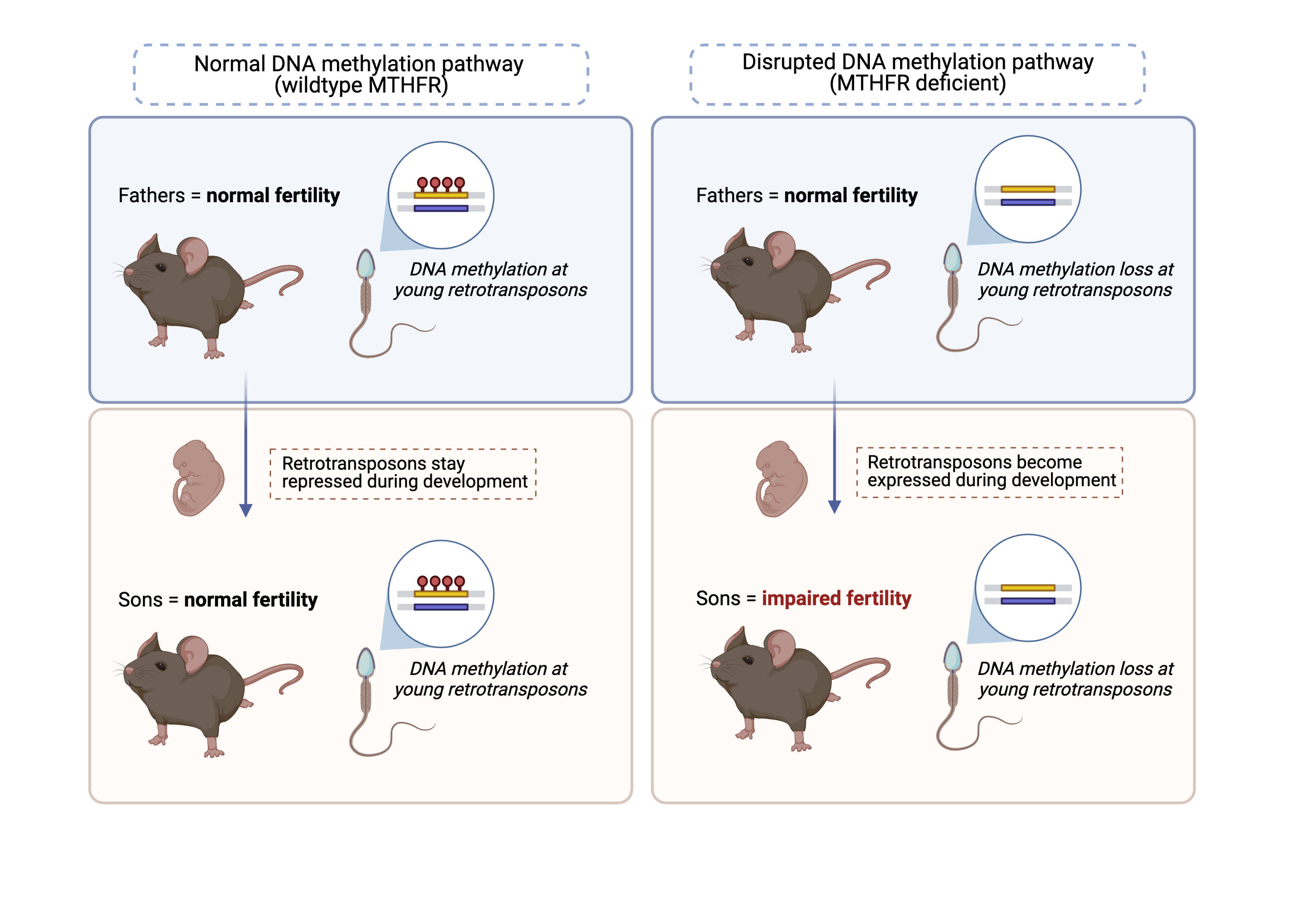
Loss of DNA methylation in sperm occurs at young retrotransposons.
Intriguingly, most of the DNA losses in sperm did not occur at coding genes, but rather in the non-coding “dark matter” of the genome. More specifically, the authors found that the sperm of both MTHFR-deficient fathers and their sons lost DNA methylation at young retrotransposons. Retrotransposons are a class of transposable element that copy-paste themselves into the genome via an RNA intermediate and can be dangerous for cells when active. Young retrotransposons are particularly harmful because they have not yet accumulated enough DNA mutations to become co-opted and beneficial to the host.
Loss of this repressive epigenetic mark at transposable elements that typically need to stay silenced during embryo development, may lead to an aberrant activation of these transposable elements, and in part explain the decline in fertility in the second generation. This study suggests that young retrotransposons may facilitate the transmission of epimutations from fathers to sons.
Learn more
- A comprehensive summary of the different classes of transposable elements: “Transposons: the jumping genes.”
- A review highlighting how transposable elements can affect genome stability but also act as genetic innovators: “Transposable elements shape the evolution of mammalian development.”
- A perspective discussing the sensitivity of transposable element to the environment: “Adaptation to global change: a transposable element-epigenetics perspective.”
We would like to thank Dr. Gurbet Karahan, Dr. Jacquetta Trasler and Dr. Sarah Kimmins from McGill University for reviewing this piece.
* Banner image: Mouse Sperm on a Counting Chamber, photo taken by Ariane Lismer

